The gene world has always fascinated us with various functions and mechanisms through which it works. Genes tell what we should do, for example, even a simple function of switching the light on or off. Won’t it be easier to diagnose or treat a disease if we knew which genes are suppressed or expressed during a particular disease progression?
So with many inventions coming up every day from around the world, we have one more innovative invention developed by Caltech scientists by using magnetic resonance imaging (MRI) to visualize gene expression in cells. This visualization technique can be used in living cells, including tumor cells, enabling the gene expression to be observed without any surgical procedures.
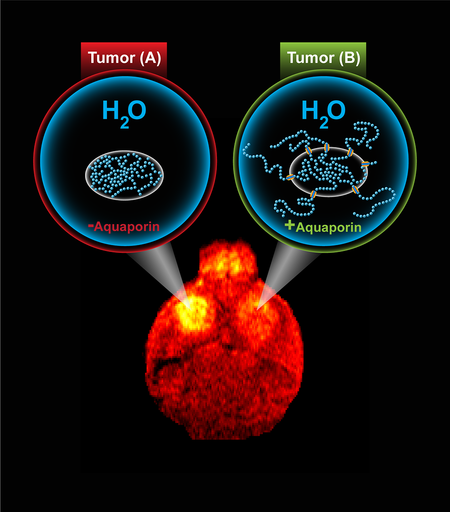
Let’s have a look how this technique works. In MRI, hydrogen atoms, which are mostly contained in fat and water molecules, are excited using a magnetic field. In turn, these excited atoms release signals that can be used to generate images of the muscle, brain, and other tissues. They can be differentiated on the basis of local chemical and physical surroundings of the water molecules.
For using water molecules, the research team used a naturally occurring protein—aquaporin. Aquaporin functions as a guard for water molecules by sitting within the membrane that envelops cells. This allows the water molecules to move in and out of the cell. Thus making aquaporin a reporter gene, which is sensitive to water molecules movements, it was linked to genes to be observed. Hence, when the gene of interest will turn on, aquaporin will be overexpressed by cells, and the cells can be observed using diffusion-weighted MRI.
The researchers successfully monitored the gene expression in mice with induced brain tumor using MRI.
Isn’t this something out-of-the-box innovation? What do you think about this technique? Feel free to share your thoughts.